# init repo notebook
!git clone https://github.com/rramosp/ppdl.git > /dev/null 2> /dev/null
!mv -n ppdl/content/init.py ppdl/content/local . 2> /dev/null
!pip install -r ppdl/content/requirements.txt > /dev/null
Distribution objects#
import numpy as np
import matplotlib.pyplot as plt
from scipy import stats
from scipy.integrate import quad
from progressbar import progressbar as pbar
from rlxutils import subplots, copy_func
import tensorflow as tf
import tensorflow_probability as tfp
tfd = tfp.distributions
tfb = tfp.bijectors
%matplotlib inline
2022-03-07 06:26:26.717202: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcudart.so.11.0'; dlerror: libcudart.so.11.0: cannot open shared object file: No such file or directory
2022-03-07 06:26:26.717231: I tensorflow/stream_executor/cuda/cudart_stub.cc:29] Ignore above cudart dlerror if you do not have a GPU set up on your machine.
import os
os.environ["CUDA_VISIBLE_DEVICES"] = "-1"
you can sample (
distribution.sample)you can compute probabilities (
distribution.log_prob)they always work with tensors
d = tfd.Normal(loc=2, scale=3)
d
2022-03-07 06:26:28.678260: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcuda.so.1'; dlerror: libcuda.so.1: cannot open shared object file: No such file or directory
2022-03-07 06:26:28.678308: W tensorflow/stream_executor/cuda/cuda_driver.cc:269] failed call to cuInit: UNKNOWN ERROR (303)
2022-03-07 06:26:28.678339: I tensorflow/stream_executor/cuda/cuda_diagnostics.cc:156] kernel driver does not appear to be running on this host (rlxyoga): /proc/driver/nvidia/version does not exist
2022-03-07 06:26:28.678764: I tensorflow/core/platform/cpu_feature_guard.cc:151] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
<tfp.distributions.Normal 'Normal' batch_shape=[] event_shape=[] dtype=float32>
s = d.sample(20)
s.shape
TensorShape([20])
s
<tf.Tensor: shape=(20,), dtype=float32, numpy=
array([ 0.84202063, -3.4651394 , 0.24238634, 4.7199984 , -1.849772 ,
0.75771403, 0.33977604, -0.0643971 , 2.414379 , 1.0733131 ,
0.32795286, -1.4055476 , 5.27031 , 3.298139 , 4.8651505 ,
4.303988 , 4.7135587 , 5.32114 , 1.4019012 , 0.2740475 ],
dtype=float32)>
d.log_prob(s)
<tf.Tensor: shape=(20,), dtype=float32, numpy=
array([-2.0920463, -3.6768703, -2.1891735, -2.4285727, -2.8409257,
-2.1032884, -2.1706812, -2.254314 , -2.0270903, -2.0652592,
-2.17287 , -2.6618707, -2.6117136, -2.1111712, -2.4736114,
-2.31246 , -2.4266286, -2.630327 , -2.0374243, -2.183046 ],
dtype=float32)>
for a larger sample
s = d.sample(10000).numpy()
xr = np.linspace(np.min(s), np.max(s), 100)
xr_probs = np.exp(d.log_prob(xr))
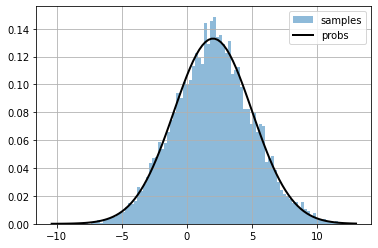
plt.hist(s, bins=100, alpha=.5, density=True, label="samples");
plt.plot(xr, xr_probs, color="black", lw=2, label="probs")
plt.grid();
plt.legend();
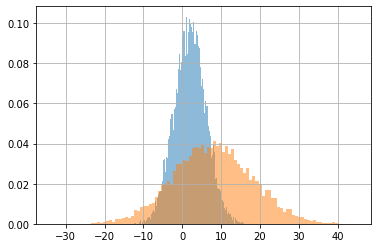
Batches of distributions#
You can simultaneously define many distributions of the same type with the shape of the parameters.
This is a batch of distributions.
All distributions in a batch are of the same type but may have different parameter values.
d = tfd.Normal(loc=[2,8], scale=[4,10])
s = d.sample(10000).numpy()
s.shape
(10000, 2)
for i in range(s.shape[1]):
plt.hist(s[:,i], alpha=.5, bins=100, density=True)
plt.grid();
The distribution configuration is stored in the distirbution object itself:
the
batch_shapethe
parameters
d.batch_shape
TensorShape([2])
d.parameters
{'loc': [2, 8],
'scale': [4, 10],
'validate_args': False,
'allow_nan_stats': True,
'name': 'Normal'}
as complex as you want#
means = np.random.randint(10, size=(2,3,4))*1. + 5
stds = np.random.randint(5, size=means.shape)*1. + 1
print(f"means\n{means}")
print(f"stds\n{stds}")
d = tfd.Normal(loc=means, scale=stds)
s = d.sample(10000).numpy()
print ("\nsample shape", s.shape)
print (f"\nthere are {np.product(s.shape[1:])} batch distributions")
means
[[[12. 13. 14. 10.]
[11. 11. 12. 5.]
[11. 5. 5. 5.]]
[[ 8. 13. 14. 7.]
[ 7. 11. 12. 10.]
[13. 7. 5. 9.]]]
stds
[[[3. 5. 2. 5.]
[5. 5. 3. 3.]
[5. 4. 5. 2.]]
[[4. 2. 4. 2.]
[4. 1. 3. 5.]
[4. 4. 3. 3.]]]
sample shape (10000, 2, 3, 4)
there are 24 batch distributions
d.batch_shape
TensorShape([2, 3, 4])
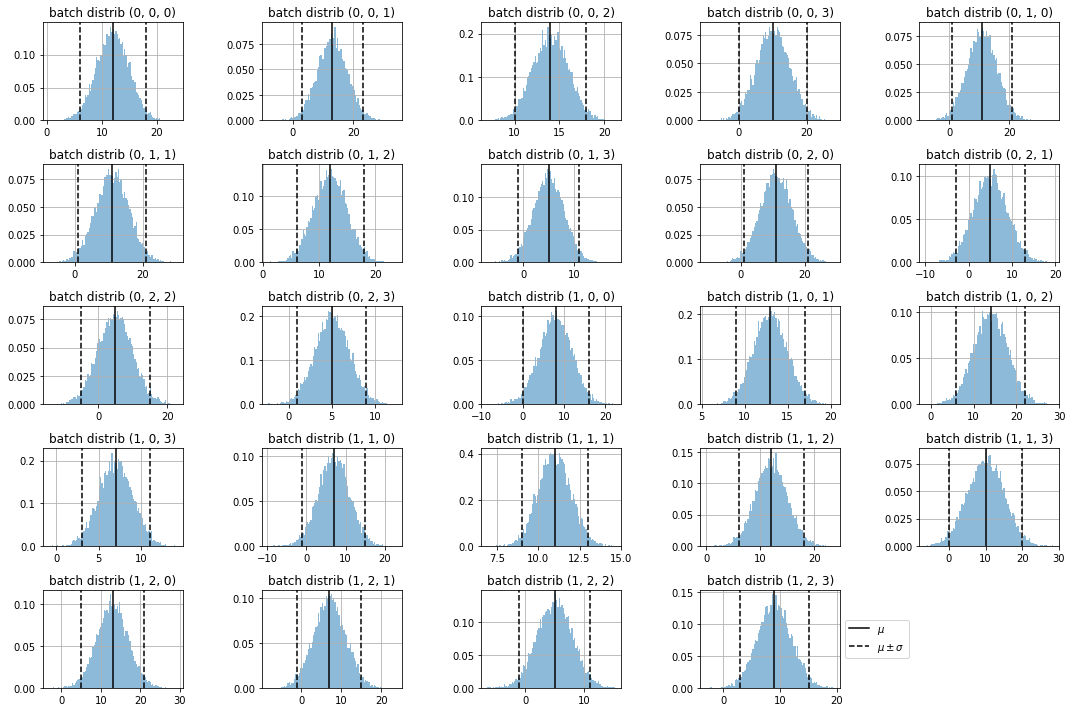
observe the samples on each batch distribution
import itertools
dims = list(itertools.product(*[range(i) for i in d.batch_shape]))
for ax, dim in subplots(dims, n_cols=5, usizey=2):
sdim = s[(..., *dim)]
plt.hist(sdim, bins=100, density=True, alpha=.5)
plt.axvline(means[dim], color="black", label="$\mu$")
plt.axvline(means[dim] + 2*stds[dim], color="black", label="$\mu \pm\sigma$", ls="--")
plt.axvline(means[dim] - 2*stds[dim], color="black", ls="--")
plt.title(f"batch distrib {dim}")
plt.grid();
if dim==dims[-1]:
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.tight_layout()
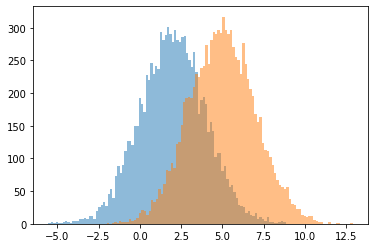
Broadcasting parameters for distribution batches#
broadcasting works between different parameters of the distributions
# a batch of two normals with the same std
d = tfd.Normal(loc=[2,5], scale=2)
s = d.sample(10000).numpy()
s.shape
(10000, 2)
d.batch_shape
TensorShape([2])
d.parameters
{'loc': [2, 5],
'scale': 2,
'validate_args': False,
'allow_nan_stats': True,
'name': 'Normal'}
for i in range(s.shape[1]):
plt.hist(s[:,i], bins=100, alpha=.5)
# a batch of two normals with the same std
means = np.r_[[[2.,5.], [8.,10.]]]
stds = np.r_[[[1.,2.]]]
print(f"means\n{means}")
print(f"stds\n{stds}")
d = tfd.Normal(loc=means, scale=stds)
s = d.sample(10000).numpy()
s.shape
means
[[ 2. 5.]
[ 8. 10.]]
stds
[[1. 2.]]
(10000, 2, 2)
d.batch_shape
TensorShape([2, 2])
dims = list(itertools.product(*[range(i) for i in s.shape[1:]]))
for dim in dims:
plt.hist(s[(...,*dim)], density=True, alpha=.5, bins=100)
plt.grid();
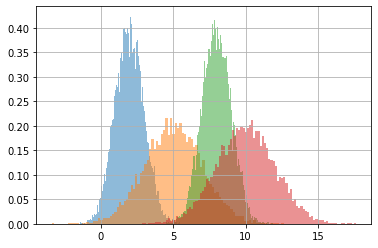
observe carefully that broadcasting is done across the dimension that matches
# a batch of two normals with the same std
print(f"means\n{means}")
print(f"stds\n{stds.T}")
d = tfd.Normal(loc=means, scale=stds.T)
s = d.sample(10000).numpy()
s.shape
means
[[ 2. 5.]
[ 8. 10.]]
stds
[[1.]
[2.]]
(10000, 2, 2)
d.batch_shape
TensorShape([2, 2])
dims = list(itertools.product(*[range(i) for i in s.shape[1:]]))
for dim in dims:
plt.hist(s[(...,*dim)], density=True, alpha=.5, bins=100)
plt.grid();
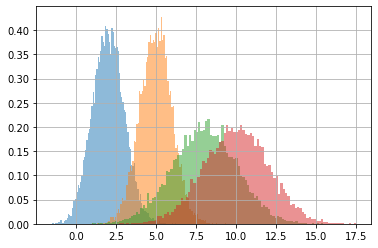
tfp distribution objects accept differentiable (learnable) parameters#
so that we can use TF variables for their parameters and then optimize
for instance, let’s fit a normal distribution to some data by maximizing the log likelihood.
assuming:
we want:
# we have some data (which actually is not generated by a Normal distribution)
x = 5*tfd.Beta(2.,5).sample(10000)+2
plt.hist(x.numpy(), bins=50, density=True, alpha=.5);
plt.axvline(np.mean(x), ls="--", color="black", alpha=.5, label="sample mean")
plt.grid(); plt.legend();
2022-03-06 05:04:36.589174: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcuda.so.1'; dlerror: libcuda.so.1: cannot open shared object file: No such file or directory
2022-03-06 05:04:36.589232: W tensorflow/stream_executor/cuda/cuda_driver.cc:269] failed call to cuInit: UNKNOWN ERROR (303)
2022-03-06 05:04:36.589273: I tensorflow/stream_executor/cuda/cuda_diagnostics.cc:156] kernel driver does not appear to be running on this host (rlxyoga): /proc/driver/nvidia/version does not exist
2022-03-06 05:04:36.590383: I tensorflow/core/platform/cpu_feature_guard.cc:151] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
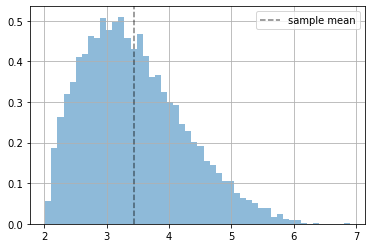
fitting a Normal distribution by hand#
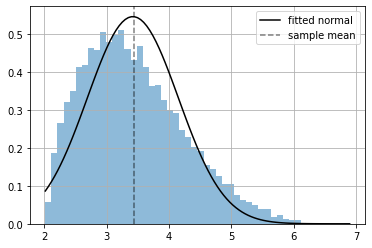
since the data actually comes from a different distribution the fitting is not so good, we are using a wrong assumption.
recall that the GradientTape must watch how the expresion you want to optimize is built, and therefore we must create the distribution object within the GradientTape
zm = tf.Variable(0, dtype=tf.float32)
zs = tf.Variable(1, dtype=tf.float32)
optimizer = tf.keras.optimizers.Adam(learning_rate=0.1)
for epoch in range(100):
with tf.GradientTape() as tape:
distribution = tfd.Normal(loc=zm, scale=tf.math.softplus(zs))
negloglik = -tf.reduce_mean(distribution.log_prob(x))
gradients = tape.gradient(negloglik, [zm, zs])
optimizer.apply_gradients(zip(gradients,[zm, zs]))
[zm, zs]
[<tf.Variable 'Variable:0' shape=() dtype=float32, numpy=3.4183526>,
<tf.Variable 'Variable:0' shape=() dtype=float32, numpy=0.0727701>]
xr = np.linspace(np.min(x), np.max(x), 100)
plt.hist(x.numpy(), bins=50, density=True, alpha=.5);
plt.plot(xr, np.exp(distribution.log_prob(xr).numpy()), color="black", label="fitted normal");
plt.axvline(np.mean(x), ls="--", color="black", alpha=.5, label="sample mean")
plt.grid(); plt.legend();
observe what would happen if we create the distribution object outside the gradient tape
zm = tf.Variable(0, dtype=tf.float32)
zs = tf.Variable(1, dtype=tf.float32)
distribution = tfd.Normal(loc=zm, scale=tf.math.softplus(zs))
optimizer = tf.keras.optimizers.Adam(learning_rate=0.1)
with tf.GradientTape() as tape:
loglik = -tf.reduce_mean(distribution.log_prob(x))
gradients = tape.gradient(loglik, [zm, zs])
optimizer.apply_gradients(zip(gradients,[zm, zs]))
WARNING:tensorflow:Gradients do not exist for variables ['Variable:0'] when minimizing the loss. If you're using `model.compile()`, did you forget to provide a `loss`argument?
<tf.Variable 'UnreadVariable' shape=() dtype=int64, numpy=1>

